

| Home Page | Overview | Site Map | Index | Appendix | Illustration | About | Contact | Update | FAQ |
 |
 |
mRNA carries genetic information from the DNA in the nucleus to the ribosomes in the cytoplasm for protein synthesis. Each gene, a segment of DNA, produces a separate mRNA molecule when a certain protein is needed in the cell, the mRNA is broken down quickly after translation. The size of an mRNA depends on the number of nucleotides in the gene. |
Figure 11-21a mRNA Transcription |
Figure 11-21b mRNA Construction |
| In the initiation stage, RNA polymerase binds to promoters and starts to unwind the DNA strands. In the elongation stage, RNA polymerase reads the DNA template stand from 3' to 5' and produces the RNA transcript from 5' to 3'. The nucleotides are always added to the 3' end of the growing RNA. In the final stage, the RNA polymerase reaches the termination site and the RNA transcription, i.e., the messenger RNA is released from the template. (Figure 11-21a.) In eukaryotes, the genes contain sections known as exons that code for proteins, are mixed with |
sections called introns that do not code for protein. A newly formed mRNA is called a pre-mRNA because it is a copy of the entire DNA template including the noncoding introns. Before the newly synthesized pre-mRNA leave the nucleus, it undergoes processing to remove the intron sections. The splicing of the pre-mRNA produces a mature, functional mRNA that leaves the nucleus to deliver the genetic information to the ribosomes for the synthesis of protein (see Figure 11-21b). |
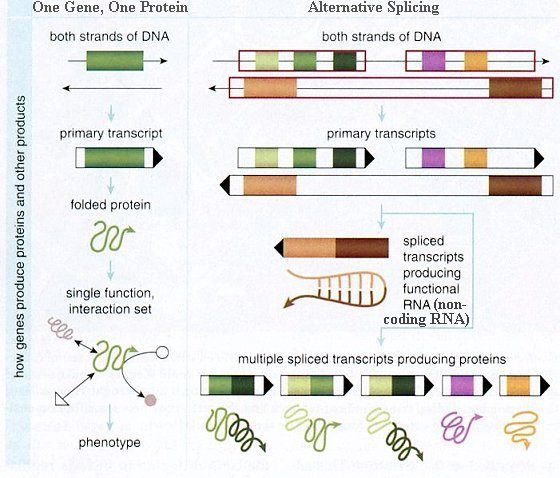 |
A 2008 genome-wide surveys of gene expression in 15 different tissues and cell lines have revealed that up to 94% of human genes generate more than one product. Only about 6% of human genes are made from a single, linear piece of DNA. Most genes are made from sections of DNA found at different locations along a strand. The data encoded in these fragments are joined together into a functional messenger RNA (mRNA) molecule that can be used as a template to generate proteins. It produces even more alternatives with the same gene assembled in different ways, sometimes leaving out a piece, for example, or including a bit of the intervening DNA sequence. This process, called alternative splicing, can produce mRNA molecules and proteins with dramatically different functions, despite being formed from the same gene (Figure 11-22). |
Figure 11-22 Alternative Splicing [view large image] |
It is found that such process happens most often in human. Thus we may have about the same number of genes as for lower animals, alternative splicing provides a way to make us more sophisticated, complex and intelligent. |
 |
 |
tRNA, the smallest of the RNA molecules, interprets the genetic information in DNA and brings specific amino acids to the ribosome for protein synthesis. Only the tRNA can translate the genetic information into amino acids for proteins. There are one or more different tRNAs for each of the 20 amino acids. The structures of the transfer RNAs are similar, consisting of 70-90 nucleotides. Hydrogen bonds between some of the complementary bases in the chain produce loops that give some double-stranded regions (See Figure 11-23). The actual structure of a tRNA has a three-dimensional L shape, (See Figure 11-24.) but it is often drawn as a cloverleaf to illustrate its features. All tRNA molecules have a 3' end with the nucleotide sequence -- ACC, which is known as the acceptor stem. An enzyme attaches an amino acid by forming an ester |
Figure 11-23 Bases in tRNA [view large image] |
Figure 11-24 Three Dimensional Structure of tRNA [view large image] |
bond with the free -- OH at the end of the acceptor stem. Each tRNA contains an anticondon, which is a series of three bases that complements the three bases on a mRNA. |
 |
 |
rRNA makes up 65% of the structural material of the ribosomes; the other 35% is protein. Ribosomes (see hybrid image§ in Figure 11-25a), which are the sites for protein synthesis, consist of two subunits, a large subunit and a small subunit. Protein synthesis requires mRNA, tRNA, amino acids, ribosomes, ATP, and various protein factors. These pieces come together at the beginning of translation, in a stage called translation initiation. Translation begins when an mRNA molecule binds to a segment of rRNA that is part of a small ribosomal subunit. The anticodon of a tRNA bearing methionine (met) bonds to the |
Figure 11-25a Ribosome |
Figure 11-25b rRNA Translation [view large image] |
initiation codon (AUG) on the mRNA. These bound structures form the initiation complex. Next, a large ribosomal subunit binds to the complex, and a tRNA bearing a second amino acid bonds between its and the second mRNA's codon. |
 |
The amino acid brough in by the first tRNA bonds with the amino acid brought in by the second tRNA, and the first tRNA detaches and floats away. The ribosome moves down the mRNA by one codon, and a third tRNA arrives, carrying another amino acid, (See Figure 11-25b.) ... and the process continues until a termination codon (UAG, UAA, UGA) is reached. Each of the three bases (the codon) in the mRNA is translated into an amino acid according to the genetic code (see Figure 11-25c). For example, an tRNA with bases CCG in the anticodon and amino acid Glycine (Gly) in the attachment site would bind to the condon GGC in the mRNA, the amino acid Glycine (Gly) would be added to the growing protein chain in the ribosome as a result of this combination. |
Figure 11-25c Genetic Code [view large image] |
 |
In the past several years, scientists have discovered a more precise and - for the purposes of research and medicine - more powerful security apparatus built into nearly all plant and animal cells. This system is called RNA interference, or RNAi, which acts like a censor. When a threatening gene is expressed, the RNAi machinery silences it by intercepting and destroying only the offender's mRNA, without disturbing the mRNAs for the other genes. RNAi also regulates the activity of normal genes during growth and development. |
Figure 11-26a RNAi |
Within the RISC, the siRNA molecule is positioned so that mRNAs can slide into it. The RISC will encounter thousands of different mRNAs that are in a typical cell at any given moment. But each siRNA of the RISC will adhere well only to a mRNA that closely complements its own nucleotide sequence. So, unlike the interferon response, the silencing complex is highly selective in choosing its target mRNAs.
When a matched mRNA finally docks onto the siRNA, an enzyme know as Slicer cuts the captured mRNA strand in two. The RISC then releases the two mRNA pieces (now rendered incapable of directing protein synthesis) and moves on. The RISC itself stays intact, free to find and cleave another mRNA. In this way, the RNAi censor uses bits of the double-stranded RNA as a "blacklist" to identify and mute corresponding mRNAs.
 |
When the RNAi machinery is not defending against attack, it apparently pitches in to help silence normal cellular genes during developmental transitions for producing disparate cell types, such as neurons and muscle cells, or different organs, such as the brain and heart. The triggers are "microRNAs" - small RNA fragments that resemble siRNAs but differ in origin. Whereas siRNAs come from the same types of genes or genomic regions that ultimately become silenced, microRNAs come from genes whose sole mission is to produce these tiny regulatory RNAs. The RNA molecule initially transcribed from a microRNA gene - the microRNA precursor - folds back on itself. With the help of Dicer, the middle section is chopped out of the microRNA, and the resulting piece typically behaves very much like an siRNA - with the important exception that it does not censor a gene with any resemblance to the one that produced it but instead censors some other gene altogether. However, this control mechanism can be nullified by attaching an inhibitor to the microRNA (Figure 11-26b). |
Figure 11-26b microRNA |