Unicellular Organisms
Genomes
See DNA Sequencing for the techniques to read the DNA text.
A gene is the part of a DNA sequence containing information about the amino acid sequence of one protein. Genes used to be studied one at a time, but with the invention of DNA sequencing machines it has become possible to consider the total DNA of an organism, usually referred to as its genome5.
The genomes of many bacteria consist of a single, circular chromosome. Human and other animal cells have linear chromosomes. An important feature of animal genomes is that much of the DNA does not code for genes. The non-coding DNA, also known as junk DNA, consists mostly of the same few sequences repeated over and over again. They are often inserted within a region of coding gene. The purpose of the noncoding DNA, if any, is not understood. As much as 97% of human DNA is noncoding. Some researches believe that they might be used as testing site for genetic mutation; other suggests that they might have a controlling function.
 |
 |
There are an estimated 20,000-25,000 human protein-coding genes (Figure 11-35a1). This number has been repeatedly revised down from initial predictions of 100,000 or more as genome sequence quality and gene finding methods have improved. The grape has an unusually high number of genes for creating flavor, e.g., more than 100 of its genes are dedicated to producing tannins (pungent taste) and terpenes (strong-smelling) - results of selective breeding by winery ?
|
| |
|
|
 |
Bacterial genomes are far more compact than eukaryotic genomes. They have very little noncoding DNA (introns). The number of genes and base pairs for some organisms are shown in Figure 11-35a. The mouse genome sequence reveals about 30,000 genes, with 99% having direct counterparts in humans. It seems to indicate that complexity is not solely determined by the number of genes, it may also be related to the regulation of these genes 7. Figure 11-35b shows that the number of chromosomes is unrelated to complexity. It could be just the error in chromosome segregation during cell |
|
division. An extra pair of chromosomes was retained by mistake. Over time, mutations would accumulate in the duplicated pair until they were so divergent that they were clearly distinct.
|
Recent research in 2003 has found that many of the non-coding genes play major roles in the health and development of plants and animals. Active forms of RNA also help to regulate a separate "epigenetic6" layer of heritable information that resides in the chromosomes but outside the DNA sequence. Lately a new kind of RNA has been discovered. Dubbed riboswitches, these long RNAs are both coding and non-coding at once. They produce protein only when activated by target chemical. These precision genetic switches have been identified from species in all five kingdoms of life. This implies that they were probably present in the last common ancestor, not long after the dawn of evolution. They may be the living relic of the RNA world 3.8 billion years ago.
The role of junk DNA becomes clearer by 2004. It is found that it may serve the function of gene regulation. The introns are not merely
 |
discarded after separating from the mRNA (the exons in the gene). Some of them are processed into MicroRNAs, which regulate the gene expression similar to some of the proteins translated by the mRNAs (see Figure 11-35c). Aside from introns, the other great source of presumed genomic junk - accounting for about 40% of the human genome - comprises transposons (a.k.a. jumping genes or transposable elements) and other repetitive elements. These sequences are widely regarded as molecular parasites that, like introns, colonized our genomes in waves at different times in evolutionary history. Evidence suggests that transposons contribute to the evolution and genomic regulation of higher organisms and may play a key role in epigenetic inheritance (the modification of genetic traits). The A-to-I (adenosine-to-inosine) editing process, in which a RNA sequence changes at a very specific site, occurs in repeat sequences call Alu elements that reside in noncoding RNA sequences. It is particularly |
| |
active in the brain, and is two orders of magnitude more widespread in humans than was previously thought. What was dismissed as junk because it was not understood may well turn out to hold the secrets to human complexity. |
By 2006 the transposable elements (TEs) are increasingly seen as major originators of genetic change, allowing populations to adapt to
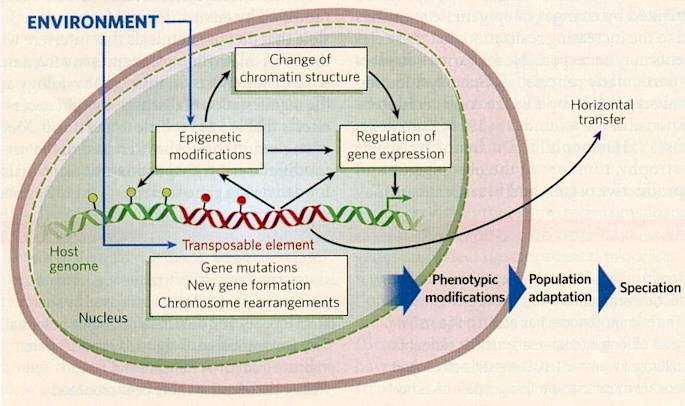 |
change and species to evolve (new phenotype produced from a genotype induced by environmental change), as shown in Figure 11-35d. They can also move between genomes of different species. Such horizontal transfer allows these elements to escape the various regulatory mechanisms imposed on them by their host genome, and to invade new genomes where they increase their copy number until new mechanisms evolve there to limit their spread. Limiting forces are also at work at the population level. These forces suggest that there is selection against the direct deleterious effects of insertions, even if these effects are small, and against the chromosomal rearrangements that frequently occur when TEs of
|
| |
the same family are present. As a result of these controlling forces, genomes contain a mixture of TEs, some of which are still active, whereas others are ancient relics that have degenerated.
|
A 2016 update confirms the role of junk DNA in controlling gene expression possibly by managing methylation (Figure 11-36a). When one piece of the junk called HACNS1 is inserted into the genomes of mouse embryos, it alter the expression of the paws and digits; this is interpreted as related to the development of opposable thumbs and walking on two legs in human. It is also found that there are 171
 |
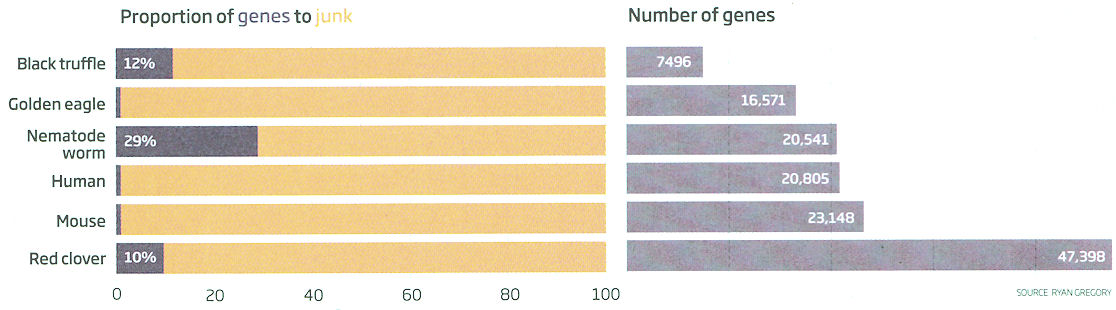 |
genes with uniquely methylation patterns in human when compare our genomes to those for chimps, gorillas and orangutans. It is suggested that there is a set of instructions for how to assemble a particular animal in junk DNA. Genetic diseases are the results of something wrong in its execution. Figure 11-36b is a 2016 update of the proportion of junk DNA in various organisms.
|
Figure 11-36a Junk DNA, Function  |
Figure 11-36b Junk DNA (2016), Proportion [view large image] |
See "You Are Junk". |
There is an unexpected application of the non-coding DNA in modern life. Since the number of repeats is highly variable among individuals, DNA profiles has been compiled to replace fingerprints as personal identification or for paternity testing.
Comparison of the genes in 100 species found only 60 genes in common to all. This number may not be enough to maintain a cell-based life form in a hypothetical "last universal common ancestor" (LUCA) as depicted in Figure 10-02b. It is possible that much of the evolutionary record has been erased from species' genomes due to gene loss as organisms adapt to new conditions and ditch redundant genetic material. The minimal gene set to produce a viable organism has been estimated initially to consist of about 250 genes; further analysis reduced the number to about 80. They are related to various functional classes such as: replication (including recombination, and repair), transcription, translation (including ribosome structure, and bio-genesis), metabolism, and cellular processes (including chaperone functions, secretion, cell division, and cell wall biogenesis).
 |
In 2006, the Craig Venter Institute started working on a minimal genome containing less than 400 genes but which nevertheless has everything it takes to sustain a free-living cell. It represents a step forward (or backward pointing to the origin of life) toward the creation of living entity from inanimated molecules. Figure 11-36c illustrates the four steps in producing the "synthetic life". In the trial run, the genome of the Mycoplasma capricolum is extracted to imitate the minimal genome (in testing the
|
| |
transplant method). The real synthesis may be just weeks or months away (as of July, 2007).
|
A progress report in January 2008 indicates that they have successfully stitched together an entire bacterial genome (about 580000 bases for the pathogenic bacterium Mycoplasma genitalium using DNA-linking enzymes) from custom-made fragments of about 5000 - 7000 bases each. It is stressed that even if a long string of DNA could be made in the lab, it could fall apart once stuck into a cell in the next step. There are many other factors that go into getting these synthetic genes to survive in cells. The ultimate plan is to produce a stripped-down version of the M. genitalium genome (the minimal genome) that might serve as a general-purpose chassis to which might be added all sorts of useful designer functions, such as genes that turn the bacteria into biological factories for making carbon-based 'green' fuels or hydrogen when fed with nutrients.
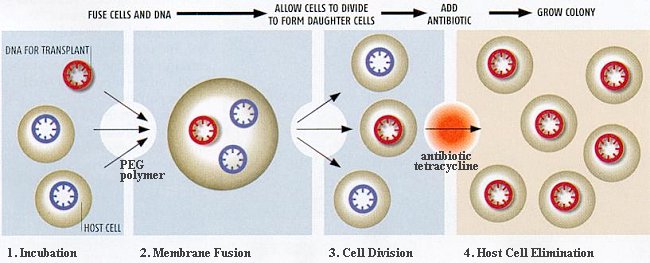
The transplanting steps (Figure 11-36c):
- Incubation - A naked circular chromosome containing the minimal genome is incubated in a rich bacterial culture.
- Membrane fusion - The solution contains a polymer called polyethylene glycol (PEG), which makes cell membranes fuse. The product sometimes has the minimal genome encapsulated inside.
- Cell division - The cell containing multiple genomes soon divides to form daughter cells.
- Elimination of host cell - The culture is then treated with the antibiotic tetracycline, which wipes out the cells containing the host genome while the cells with the minimal genome survive and grow.
The Craig Venter team announced in May 2010 that they have successfully recreated life with the following steps:
 |
1. They bought 1000 1080-base lifeless DNA sequences from a company. The whole M. mycoides bacterial genome is contained in this pool.
2. To facilitate their assembly in the correct order, the ends of each sequence had 80 bases overlapped with its neighbors. This is the "watermark" to identify the assembled genome as synthetic. In addition, 4 of the ordered DNA sequences code the e-mail address of the team member to double check the identity.
3. Yeast is used to assemble the synthetic DNA in stages - stitching them together in the order of 10,000-base first, then 100,000 and finally the complete genome.
4. This bacterial genome is used to replace the genome of another bacterium called M. capricolum. After
|
|
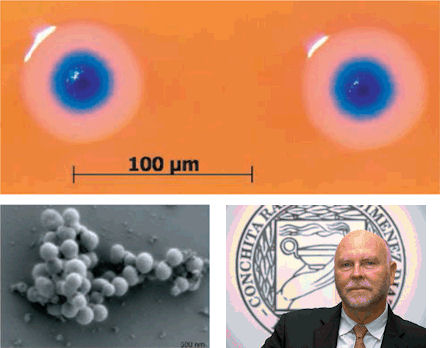
months of trying, they eventually found one morning that a blue colony of bacteria had rapidly grown on a lab plate (Figure 11-36d, with the self-replicating bacteria in an electron micrograph, and a portrait of Dr. J. Craig). |
 |
Since the effort required a lot of helps from other living organisms, this experiment actually demonstrated only partially that some lifeless macro-molecules can be transformed to a living entity, which can replicate itself. Such event would not happen on Earth 4 billion years ago when it was totally lifeless. Anyway, it would be a successful venture for Dr. J. Craig and company as their purpose is to produce fuel, oil dispersant, garbage disposal, ... etc via a particular sequence of DNA (Figure 11-36d1). |
Figure 11-36d1 Applications of Synthetic Genomes  |
|
The artificial cell created in 2010 contains an almost exact copy of the wild-type M. mycoides genome, except that the DNA sequence has been re-assembled from pieces of the original with the addition of a few watermark for identification. It was dubbed as JCVIsyn1.0 containing about 1 million base pairs and 900 genes. Since then, improvements in technology and methodology have enabled the creation of a minimal genome of 531,560 base pairs and 473 genes. This is the JCVIsyn3.0 heralded in a March 24, 2016 proclamation to the world. Followings are a summary of the synthetic process (the Design-Build-Test (DBT) cycle) and some specific properties of the artificial cell.
 |
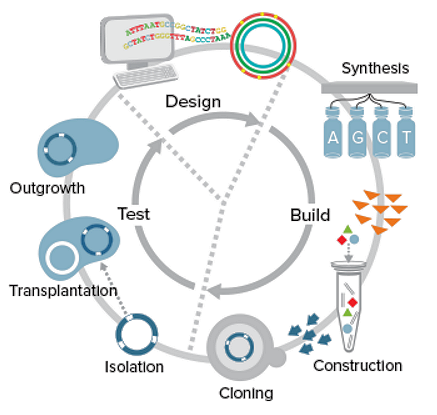 |

|
Figure 11-36d2 Genome Synthesis 
|
|
|
Design-Build-Test (DBT) cycle :
- Original Material - Oligonucleotides (DNA segments) from syn1.0 genome were supplied by a commercial company.
- DNA Sequence Design - Design software is used to determine specific of the 1.4-kbp fragments for later assembly including overlapping ends, vector sequence (for DNA insertion), restriction sites (the cutting point), and watermark (for identification), ...
- Intermediate Assembly - The 1.4-kbp fragments are transformed into E. coli and 5 of each were combined into 7-kbp cassettes (mobile genetic element). The whole genome are divided into eight different segments to facilitate verification.
- Assembly in Yeast - The eight different types of cassettes are introduced into yeast to be sequence verified and the complete genome is finally generated in the form of centromeric plasmid (Figure 11-36d2) containing various combination between the cassettes and portion of the original syn1.0 genome (see Figure 11-36d4, column 4). The four color circles in Figure 11-362 illustrate the steps to assemble the synthetic genome from smaller pieces of oligonucleotide.
- Transplantation - The synthetic genomes were transplanted out of the yeast into an M. capricolum recipient cell for testing in viability and other phenotypic traits. This marks the completion of one DBT cycle as shown in Figure 11-36d3.
- Cycles - It required three rounds of Reduced Genome Design (RGD) to arrive at the minimal synthetic genome as shown in Figure 11-36d4. The efforts are mostly on deletions of un-necessary genes. The final product is viable as well as having a acceptable growth rate, i.e., the doubling time td
 100 minutes.
100 minutes.
Characteristics of JCVIsyn3.0 :
- The minimal synthetic genome contains 473 genes - about 1/2 of those in its predecessor syn1.0. The minimum number of genes for a viable organism actually depends on the environment. The minimal 473 genes are for those cells living in nutrient-rich petri dish and thus some metabolic genes are not required. The number should increase in harsh condition. As observed by Dr. Venter himself : "... there is no such thing as a true minimal genome."
- Figure 11-36d5 shows the portion of the genes in four functional groups. The function of 149 of these are unknown even many of them are found in other life form. They are probably molecular switches to turn on and off gene expression. But it reveals that we understand only a little part of life's symphony so far.
- The purpose of creating syn3.0 is to assess the minimum requirment for a viable genome, from which other genes can be added to
 |
 |
manufacture useful products. Thus, the housing for the genome provided by another organism is just a simplification of the process, which is not for the understanding of the "origin of life". Thus, it should not be concluded that cell membranes were created before the genes in the "chicken-or-egg" debate. Anyway, the very popular CRISPR gene-editing technique is suitable to make a few changes in the gene, while the whole genome approach is necessary for designing something new.
|
|
|
- Figure 11-36d6 shows a cluster of the cells containing the syn3.0 genome. The colonies are morphologically similar to those of JCVI-syn1.0, and appears to be polymorphic (multiple form) when examined microscopically.
|
See the original article : "Design and Synthesis of a Minimal Bacterial Genome".
More recent study (of the genomes of Archaea, bacteria, fungi, plants, and animals) expands the number of "immortal" genes to about 500. These genes have survived through an immense time of about 2 billion years and life will continue to depend upon this core set of genes as it evolves in the future. It is noted that most of the similarities between archaea and eukaryotes were in so-called informational genes whose products dealt with the copying and decoding of DNA; while most of the similarities between eukaryptes and bacteria were in operational genes involved in the metabolism of various nutrients and basic cellular materials. It appeared as though the eukaryotes got their "brains" (informational genes) from one parent, and their "looks" (operational genes) from another (see "ring of life"). The two types of early life (the archaea, and bacteria) have completely different cell membrane. It seems that the subsets of LUCA broke away on two separate occasions acquiring the characteristics of two differnt types .
The creation of complex objects, whether houses or horses, demands two kinds of specifications: one for the components and one for the system that guides their assembly. The component molecules that make up different organisms are fundamentally alike: around 99% of the proteins in humans have recognizable equivalents in mice, and vice versa; many of those proteins are also conserved in other animals, and those involved in basic cellular processes are conserved in all eukaryotes. So it must be the architectural information that accounts for the diversity of animals. Since the amount of regulation increases as a nonlinear function of complex and protein regulation has its
 |
limitation, it is suggested that the rise of multicellular organisms over the past billion years was a consequence of the transition to a new control mechanism based largely on RNA regulatory signals from the junk DNAs. The evolution of complexity (in term of new regulatory system) helps to explain the phenomenon of the Cambrian explosion about 52.5 million years ago, when invertebrate animals evolved, seemingly abruptly, from much simpler life (see Figure 11-36e). Beside the architectural plans, there is the requirement of energy to do the actual construction. This is available only when a bacterium ended up inside an archaeon about a billion years ago. The union broke the energy barrier and allowed the building of complex structure in multicellular organisms. |
Figure 11-36e Evolution of Complexity
[view large image]
|
|
Another study shows that when two large non-coding "gene deserts" were removed from the mouse genome, not only were the resulting mice viable, but their morphology, reproductive fitness, growth and longevity were indistinguishable from normal litter mates. Though some of the deleted sequences may encode functions not yet identified, the good health of these mice does suggest that there is disposable DNA in the genomes of mammals. This finding is in contrary to those just mentioned above. A possible explanation for the contradiction could be that there are so many copies of the non-coding sequence, deletion of one or two million such base pairs does not affect the biological functions.
Genes reside in the coding regions of the DNA. Normally, there are two copies of the same gene - one from each parent (see Top of Figure 11-36f). It is found that missing or extra copy of gene can cause disease in people as well as animals (see bottom of Figure 11-36f). Gene copy number variants can alter the amount of protein produced. Cell with three or more copies of a gene will tend to produce
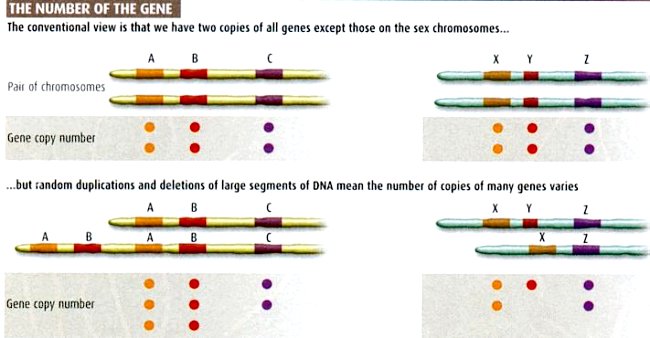 |
more of the protein the gene codes for than cells with the standard two copies. Because women have two copies of the X chromosome, most of the genes on one of the Xs are switched off to avoid double-dosing on these proteins compared with men, and usually only one of the alleles is expressed in the gene pair. However, not all the gene copy number variants would cause problems; it seems that many biologically important effects will only become apparent under certain conditions or at certain times in a person's life. Meanwhile, variants in gene copy number have been linked to autism, schizophrenia, bipolar disorder, Parkinson's, a kidney disease and a rare, inherited form of early-onset Alzheimer's disease. |
| |
|
By 2008, it has been widely recognized that human genome is very similar to other vertebrates with counterpart in them for at least 99% of all our genes. It is known that fewer than 10% of all genes are devoted to the construction and patterning of animal bodies during their
 |
development from fertilized egg to adult. The rest are involved in the everyday tasks of cells within various organs and tissues. The discovery that body-building proteins are even more alike on average than other proteins seem to contradict with the diversification of anatomical forms. It turns out that certain noncoding DNA sequences play a critical part in directing when and where to express a particular gene. They are components of "genetic switches" that turn genes on or off at the right time and place in fetal development and growth. The transcription factors (DNA-binding proteins) recognize those DNA sequences known as enhancers. There can be many enhancers for a given gene. Each enhancer specifies a particular trait of an animal. Figure 11-36g shows just two enhancer sites that control the colour in the wing and abdomen of the fruit fly. This is a versatile way of making different anatomical features without changing the gene itself. Other examples |
|
include the disappearanceof pelvic fin in the shallow-water stickleback, and the removal of a red blood cell receptor in West African population making them vulnerable to the malarial parasite. The next level of inquiry would be to understand the mechanism to turn on/off the enhancer at the right place and correct time. |
5The human genome selected the most common alleles over a number of individuals or from just one person. DNA sequence variations among individuals do occur, it is called polymorphisms. Alleles are detectable variations occurring at a single genetic locus (location). Where allelic variation is frequently found (say that at least 10% of chromosomes have an allele other than the most commonly occurring one) one refers to "a polymorphism". If variation is rare one is more likely to speak of "a mutation". SNP (Single Nucleotide Polymorphisms) refers to variation of just one nucleotide. The SNP consortium (TSC) is a public/private collaboration that has to date discovered and characterized nearly 1.8 million SNPs, which are important in tracing the evolution of the human race and controlling human diseases.
6Epigenetics is the study of heritable changes in gene function that occur without a change in the DNA sequence. Epigenetic mechanisms includes histone modification, DNA methylation (replacing H with CH3), and RNA interference. DNA methylation is to add a methyl group to the DNA - frequently to the base cytosine when it is immediately followed by guanine. The methyl group can be sensed by proteins that turn gene expression on or off through regulating chromatin structure. Histone modification involves the chemical tags attached to the "tails" of the histones. There are more than twenty different tags, or certain combinations of them, that can either give rise to relaxed chromatin, which allows the assembly of transcription factors and transcription by RNA polymerase, or produce the opposite effect. If the DNA sequence of the genome is like the musical score in a song, then the epigenome is like the musical notations that show how the notes of the melody should be played. The sequence of the human genome is the same in all our cells, whereas the epigenome differs from tissue to tissue, and changes in response to the cell's environment. Their effects in gene inactivation and activation are increasingly understood to be very important in phenotype transmission and embryonic development.
7The birth of CC (a.k.a. Copy Cat), the cloned cat, shows that the characteristics of the clone can be very different from its genetic parent. Recent work in pig cloning found that some attributes - such as the levels of albumin and calcium in blood - varied less in clones than in a control group of naturally bred pigs. Yet a surprising variety of other traits - including blood glucose and globulins, hair type, number of teats and weight - fluctuate as much in clones as in controls. These characteristics, like the pattern of CC's coat, are influenced by environmental factors and "epigenetic" controls that affect gene expression.
.














 100 minutes.
100 minutes. 



