
| Home Page | Overview | Site Map | Index | Appendix | Illustration | Preface | Contact | Update | FAQ |
 |
more about the concepts of self-assembly, self-organization and the phenomena of emergence (from the prebiotic soup, Figure 01). The explanations are all based on the science of physics and chemistry although ultimately life could be better described at the level of biology. Giving enough time, life is inevitable provided the condition is right. For example, the hydrogen bond so important for maintaining the structure of life will be disrupted by thermal agitation at few tens |
Figure 01 Origin of Life |
of degrees above room temperature; and the structure becomes frozen if temperature is low enough for the van der Waals force to take over. |
 ). Non-covalent interactions such as hydrogen bonding, and van der Waals forces then serve as the glue that holds the 2 molecules together. Examples include the interactions between ligands and receptors, the docking of antibodies on antigen molecules, the binding of DNA/polymerases, and the instant self-assembly of the ribosome components in presence of mRNA.
). Non-covalent interactions such as hydrogen bonding, and van der Waals forces then serve as the glue that holds the 2 molecules together. Examples include the interactions between ligands and receptors, the docking of antibodies on antigen molecules, the binding of DNA/polymerases, and the instant self-assembly of the ribosome components in presence of mRNA. |
|
Figure 02 Self-assembly |
Figure 02 shows some examples of self-assembly from peptide components to nano-structures. The alphabet in there represent the individual amino acid. |
 |
more elementary, which can be traced back to the electromagnetic force; the interaction in self-organization involves the kind of action that affects many facets. The size of the resulting structure from self-assembly is mostly mesoscopic, but it can be considerably larger for self-organization. Figure 03 illustrates the continuum of open thermodynamic systems from ordered, near-to-chaotic, to far-from-equilibrium states. As energy consumption increases, systems move further from equilibrium and pass through a phase transition between order and chaos. Complex systems such as proto-ribosome exist in this phase transition. |
Figure 03 Self-organization |
The diagram also shows the assembly and self-organizational activity of the proto-ribosome under the sun (for energy infusion). It is supposed to be the ancestor of the modern ribosome making protein from genetic codes. |
 |
macroscopic correspondence offer an example of emergent phenomena without self-organization - it is a change of perspective from individual molecules to thermodynamics without any change in the entropy of the system. The example of slime moulds life cycle is for |
Figure 05 Examples of Auto-systems [view large image] |
the case of simultaneous occurrence of self-organization and emergent phenomena (Figure 05). |
 |
The self-organization and emergent phenomena occur simultaneously when the slime moulds (see more in "Slime Moulds Life Cycle") run out of food. As shown in Figure 06, The parts are the individual amoebae, the interaction is the adhesive molecules, the energy is supplied by their favorite food - the bacteria and yeast, and the emergent phenomena is the moving slug and fruiting body. |
Figure 06 Slime Mould |
 |
Most theories on the origin of life adopt a "bottom-up" approach from simple to complex and from unknown to known. Another method is to trace the evolutionary path from a known point at present. The path is narrowing down by observing a conversed organelle called ribosome inside all life including bacteria and human. Although difference exists on the ribosome across different species, there is a core structure inside that is present in all of them. The following is an attempt to reach the origin by such scenario from "The Modern Day Ribosome" (structure and function) to "History of Ribosome" (looking backward to LUCA). |
Figure 07 RNA World |
Figure 07 portrays one of the bottom-up theories - the hypothetical "RNA World" in a evolutionary sequence from some nucleotides ~ 4.5 Gyr ago to LUCA (Last Universal Common Ancestor) ~ one billion years later. See relationship to the "RNA-Protein World" in Step C. |
 |
The generation of peptide chain occurs at the active site, where the rRNA in the LSU acts like an enzyme (ribozyme, aka peptidyl transferase) to add another amino acid from the tRNA at A-site to the growing peptide chain at P-site (Figure 10). The SSU is the decoding site. 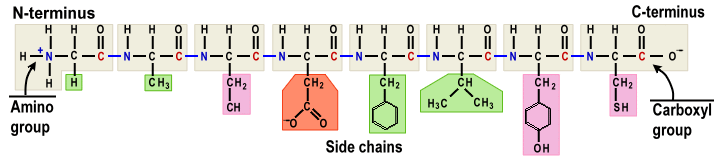  [peptide chain, 08a]. [peptide chain, 08a].In chemical formula : |
Figure 08 Genetic Code / Amino Acids / Peptide Chain [view large image] |
(peptide chain)n + ribosome + mRNA + (tRNA + amino acid) + ATP + H2O 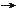
(peptide chain)n+1 + ribosome + mRNA + tRNA + AMP + PPi + 0.64 ev |
 |
 |
 |
The aggregate also recruits proteins called elongation factors (EF) to transfer the tRNA into the ribosome at A-site and to push the mRNA to the P-site for adding another amino acid to the growing peptide chain. The action is self explanatory by Figures 10, 11; while the video in |
Figure 09 Ribosome Structure |
Figure 10 Ribosome and tRNA in Action |
Figure 11 mRNA |
Figure 11 further articulates the processing sequence which moves the whole thing to an ER at the end. |
 |
 |
back to form double helix) labeled by numerals from 7 to 10 (see Figure 13,A) with a total length of 92 nt (nucleotide). The difference to this basic form in other species occurs mostly from the extension of helix 9 (shown in different color in different species). Figure 13,B is the secondary structure, 13,C the 3D super-imposition for H. sapiens, 13,D shows the region of insertion (fingerprint). According to this sequence of variations, the common core can be considered as the basic structure of the SSU rRNA soon after the emergence of life. There is similarly a basic core for the LSU rRNA. |
Figure 12 Ribosome Assembly |
Figure 13 Ribosome SSU Evolution |
  see 3-D Pol III and a video on "cryo-EM" (for making such image). see 3-D Pol III and a video on "cryo-EM" (for making such image). |
 |
Figure 14 Ribosome Evolution |
 (metal + + 1 free e-)] and the folding protects them against chemical degradation.
(metal + + 1 free e-)] and the folding protects them against chemical degradation. |
|
Figure 14b Modern Ribosome |
The Svedberg unit (such as 50S) offers a measure depending on the particle's size, mass, density, and shape indirectly based on its sedimentation rate under acceleration. |
 |
|
Figure 14d rRNA Subunits |
extant ribosome, and in consistent with further researches on primitive ribosome as shown below. |
 |
|
Figure 14e |
 |
 |
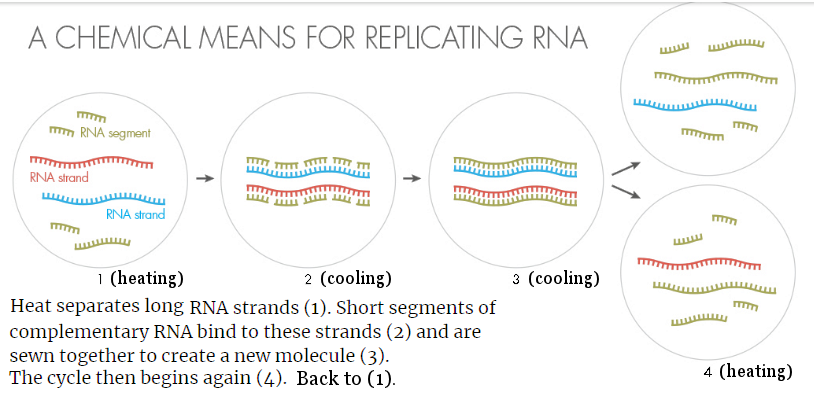
The model above is essentially the theory of RNA world looking backward. It postulates the RNA to perform both the replicative and catalytic functions. While catalyst can be made from RNA folding, RNA self-replication is difficult to create in laboratory. One suggestion is by ribozyme catalyses as shown in Figure 07, |
Figure 15 RNA Replication by Heating and Cooling |
Figure 16 Protein/RNA World [view large image] |
Another theory considers heat separation of the RNA minihelix and piecemeal assembly as shown in Figure 15. |
 |
role of aaRS is only implied in the formation of the proto-CCA (Figure 10). The RNA World follows the evolution of ribosome to arrive at the ancestral rRNA. While the Protein/RNA World as portrayed in Figure 16 emphasizes the role played by aaRS, which bears evidence of ancestral protein (enzyme) in the very beginning of the evolution of life. These two theories are complementary to each other. Evolution on both ribosome and aaRS should be considered in any future investigation into the "Origin of Life". For example, there would be no translation by the ribosome without the tRNA charged by the aaRS. See "Origin of Translation" for more info on aaRS in action. |
Figure 17 aaRS in Action |
BTW, most cells contain 20 different aaRS enzymes, one for each amino acid. The editing function of the aaRS as indicated in Figure 17 is to ensure high fidelity of tRNA charging. |
 |
These RNA segments correspond to the charged/un-charged modern day tRNA but carrying only 1 nucleoside instead of 3 (the codon carrying the genetic code in modern version). This step corresponds to the transfer of amino acides from the 1st tRNA to the next one within the ribosome (see Figure 10). This step corresponds to the advance of mRNA fetching the next tRNA to the ribosome (Figure 10). |
Figure 17b RNA/Protein World [view large image] |
then returns to step 3 and keeps on repeating, growing a short chain of amino acids — a mini-protein called a peptide — that grew while still attached to the RNA - the process is uncontrollable. This one corresponds to the elongation after processing each tRNA in the ribosome (Figure 10) - the process is regulated by mRNA in the ribosome. |

 |
 |
|
Figure 17c PTC |
Figure 17d Evolution of Ribosome |
Figure 17d shows the evolution of ribosome in quaternary structures. |
 |
  [structure of peptide, side chains (residues) in colors]. [structure of peptide, side chains (residues) in colors]. |
Figure 17e Genetic Code and |
In the translational process, the amino acid is joined by its carboxyl group to the 3' OH of the tRNA (see Figure 17f), while the amino group at the other end (of the incoming amino acid) is added to the growing peptide chain with the release of a water molecule (H2O). |
 |
structure of rRNA consists of stem-loop (ladder-bubble) configuration looking somewhat like hairpins, formed by base pairing between complementary nucleotides. These structures are stabilized by hydrogen bonds between the base pairs, from which the shape is predicted. The pairing is absent inside the loop/bubble leaving it exposed to interact with external molecules such as the codon in modern tRNA (see Figures 17f, and 17g). |
Figure 17f Modern tRNA |
The structure is revealed by various experimental techniques such as X-ray crystallography or cryo-electron microscopy. |
 |
is essential for positioning the P-site tRNA during peptidyl transfer, and its mutation severely affects the PTC activity.
|
Figure 17h Ribosomal, Details and Innards [view large image] |
See "The Peptidyl Transferase Center: a Window to the Past" for a lot more information on functional implications (TABLE 1), the position of the nucleotides in secondary structure (FIG 1), ... |
 |
Initially, the primordial soup contained single nucleotides, short RNA segments, and RNA chains of significant size (~ 50-90 nucleotides), which survived since they acquired a stable conformation. These surviving ancient RNA chains could have been the ancestors of RNA chains now referred to as proto-ribosome which underwent dimerization (formation of a dimer molecule form 2 monomer units) subsequently, and thus constructing a moiety resembling the symmetrical region in the extant ribosome. Figure 17i shows the proto-ribosome formation from a RNA precursor, undergoing simple catalytic peptidyl transferase activity. The substrates (substances on which an enzyme acts) could be one of the amino acids without codon association. |
Figure 17i Proto-Ribosome, Orgin [view large image] |
See "The Proto-Ribosome: an ancient nano-machine for peptide bond formation". |
 |
|
Figure 17j Pre-tRNA, Model [view large image] |
(see Figure 17e, "Quick Guide - Genetic Code", and "Errors in Translational Decoding"). |

 |
there is a suitable entry point at the surface of some vulnerable cell (see COVID-19). Figure 18 shows the replication process for the RNA virus. After entering the cell by endocytosis, the virus becomes uncoated. The DNA then replicates more of its kind and simultaneously making new coating proteins. These parts assemble to form more viruses, which exit from the host to infect more cells. One of the theories proposes that viruses originated and coevolved along with the most primitive cells that first contained self-replicating abilities. Irrespective of the various hypotheses, the fact that the viral parts re-pack themselves, constitutes the strongest support for the role of self-organization and emergent phenomena in the origin of life. This case involves the self-assembly of macromolecules such as RNA, and proteins (the parts) inside the host. The self-organization occurs within the host at its expense. Finally, a new virus (a coherent whole entity) appears with emergent property which can infect another cell. This is everyday occurrence in modern time. It seems that the virus stops further evolution at a very primitive stage, while the cell continuous on with further development to very complicated organism. The coevolution stopped at a point when the virus adopted its parasitic existence with no need for much improvement. |
Figure 18 RNA Virus |
See "coevolution scenario". The followings provides further details on RNA virus replication in 7 steps (Figure 18) : |
 |
|
Figure 19a Virus Self-assembly |
 |
|
Figure 19b Virus Habitats |
The number of virus groups in various environments is inscribed on the perimeter of the circle in Figure 19b. |
 |
There are many interpretations on the origin of viruses. A single evolutionary sequence can be constructed by considering the progressive change of their characteristics such as the length of genome, physical size etc. (Figure 19c,c). |
Figure 19c Virus Evolution - |
Following is an attempt to link the various kind of microscpoic organisms together in an evolutionary sequence (aka Co-evolution Theory). |
 - These are simplest organisms consisting only of a short chain of naked RNA containing 240 - 375 base pairs (bp). There is no capsid to house the genetic material. They should be around even earlier than RNA virus in the "Pre-RNA" era about 4 billion years ago (see Figure 19d).
- These are simplest organisms consisting only of a short chain of naked RNA containing 240 - 375 base pairs (bp). There is no capsid to house the genetic material. They should be around even earlier than RNA virus in the "Pre-RNA" era about 4 billion years ago (see Figure 19d). |
 |
| Figure 19d Transition to Life [view large image] | Figure 19e Complexity of Life  |
 |
 |
This kind of processing is very similar to the functions of the various polymerases (Pol's) and ribosome. Thus, the RNA virus misses only a few steps before coming alive. The crucial missing link is the absence of ATP. There is no way to attain a non-equilibrium state for living entity without the continuous supply of energy (see "Energy Supplier for Life"). Table 01 below compares the viral replication with similar modern processes in living organisms (see a pictorial illustration in Figure 19g). |
Figure 19f Virus Infection |
Figure 19g Viral Replication, Pol II, I, III, mRNA Translation |
| Process | Template | Output | Composition | Function |
|---|---|---|---|---|
| Viral Replication | RNA | RNA | Complex of 6 enzymes | Replicate viral RNA |
| RNA Polymerases II | DNA | RNA | Complex of 12 protein subunits including TBP and TRF2 proteins |
Transcribe DNA into precursors of mRNA |
| RNA Polymerases I | DNA | rRNA | Enzyme of 14 protein subunits | Transcribe DNA into rRNA |
| RNA Polymerases III | DNA | tRNA | Complex of many transcription factors | Transcribe DNA into tRNA |
| Ribosomal Translaton | mRNA | Protein | 65% rRNA and 35% proteins | Translate mRNA to protein |

 |
 |
The electron configuration of the normal carbon atom has 2 electrons in energy level 2S and 2P respectively. By supplying about 2 ev to a carbon atom, the 4 electrons in the 2S and 2P states are rearranged to the SP3 state (Figure 20). The four electrons in the SP3 state form the tetrahedral arrangement (Figure 21) of orbitals (probability distribution of electrons), which can form stable covalent bonds with other atoms. |
Figure 20 [view large image] | Figure 21 Tetrahedra |
It is no accident that photosynthesis supplies 36 ATPs each carrying ~ 0.32 ev to synthesize 1 glucose molecule.  |
 |
The stand alone SP3 excited state has a half-life about 10-15 sec, the tetrahedra would break down and return to equilibrium (see Figure 22) in absence of energy infusion in the form of Sun light to have it recycled (see "Photosynthesis - Respiration"). The stability of the SP3 state in organic molecules like carbohydrates (glucose) is maintained by the strength of the covalent bonds and is not subject to spontaneous de-excitation for the individual carbon atom (Figure 21a). The energy stored in SP3 is released with the break up of the covalent bonds. |
Figure 21a SP3 State in Glucose |
The 2 ev energy released by de-excitation of SP3 is distributed in 6 ATP ~ 0.32 ev each.
ChatGPT provides further details (in italic) : |
 |
structures are living systems, non-life examples include convection, hurricanes, the Solar system, and galaxies, ... Living system can form only when the dissipative structure begins to perform work. As the hybrid orbitals of the tetrahedral configuration do not exist in an isolated atom, but arise while the carbon atom is interacting with others to form a molecule, it will dissolve and return to an equilibrium (lowest energy) state once the input of free energy ceases causing the removal of the associated constituents. |
Figure 22 Dissipative Process |
BTW, life is optimized to an near-equilibrium state to make it a highly efficient process in using free energy. |
 |
as shown in Figure 22. Such requirement is fulfilled by re-transmission of lower frequency radiation back to the cold dark space as illustrated in Figure 23. The process re-emits more photons (at lower frequency) back to the space. By Boltzmann's definition of entropy, it has the effect of enlarging the phase space volume and hence returning more entropy to space than receiving from the sun in accordance with the second law of thermodynamics. |
Figure 23 Entropy and Life |
Ultimately, it is the stars which generate the carbon atoms (see "Origin of Elements"), and the Sun which supplies energy to all life on Earth. |
 |
 |
molecules such as iodine, oil, or grease do not dissolve in water because water is polar. The polarities of a solute and a solvent must be similar in order to form a solution. It is found as early as the 1800s that whenever a substance is dissolved in a liquid the vapor pressure (the pressure at which the rate of evaporation is equal to the rate of condensation) of the solvent is lowered. Such behaviour has the effect of rising the boiling point and lowering the freezing point of the solution. It also produces osmosis, which is the flow of a solvent, |
Figure 24 Solution |
Figure 25 Types of Solution [view large image] |
usually water, through a semipermeable membrane into a solution of higher solute concentration, i.e., diluting the solution. |
 |
 |
is about 10 times weaker than the covalent and ionic bonds. It is important in fixing properties such as solubilities, melting points, boiling points, and in determining the form and stability of crystalline structures. Molecules such as water carrying hydrogen bonds are called polar molecules. They play a crucial role in biological systems. |
Figure 26 Hydrogen Bond |
Figure 27 Water [view large image] |
 |
While unicelluar organisms can acquire nutrients and expel waste directly in aquatic environment, they could not survive on dry surfaces with a humidity of less than 10%. The same situation is applicable to the cells in multicelluar organisms which solve the problem with the "Internal Sea" in the form of Interstitial Fluid (ISF). Figure 28 also shows that the fluid composition inside the cell (Intracellular Fluid, ICF) is quite different from sea water as the constituents are altered by the metabolic process of life. |
Figure 28 Sea Water and Body Fluids, |
See "Interstitial Fluid" for more detail. |
 |
 |
to speed up a reaction is to lower the activation energy Ea. This can be done by adding a catalyst, which speeds up the reaction but is not itself changed or used up. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Most are proteins, although a few are RNA molecules (ribozymes). The catalyzing function comes from the unique 3-dimensional structures. |
Figure 29 Reaction Rate for Simple Molecules [view large image] |
Figure 30 Enzyme |
Enzymes are classified according to the chemical reactions they catalyze. See "Enzyme Commission (EC) number". |
 |
See 2024 article "A shallow lake in Canada could point to the origin of life on Earth" |
Figure 31 Prebiotic World |
about a phosphate-rich lake in Canada.
Anyway, all such short polymers can be produced at moderate temperature range and sufficient concentration without the assistant of enzyme (Figure 29). |
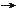 (AMP + energy) to establish the C-C, C-N, C-O, or C-S bonds (see Figure 17 and Figure 31,2).
(AMP + energy) to establish the C-C, C-N, C-O, or C-S bonds (see Figure 17 and Figure 31,2).

 |
different types of polymerase; for example, the RNA polymerase is composed of a dozen different proteins. Together, they form a machine that surrounds a DNA strands, unwinds them, and builds an RNA strand based on the information held inside the DNA. Once the enzyme gets started, RNA polymerase marches along the DNA copying RNA strands thousands of nucleotides long (Figure 32). |
Figure 32 RNA Polymerase in Action [view large image] |
Another example is the DNA replication expressed by the formula : deoxynucleoside triphosphate + DNAn + DNA-pol  diphosphate + DNAn+1 + DNA-pol diphosphate + DNAn+1 + DNA-pol |
 Charged-tRNA + AMP + PPi + aaRS.
Charged-tRNA + AMP + PPi + aaRS. |
kind, and so on, ... with the enzyme along the way to facilitate the process by lowering the activation energy. (see Figures 29 and 30 respectively for the relationship between reaction rate and concentration and the effect of enzyme in lowering the activation energy).
|
Figure 33 Binding Energy |
For example, the binding energy of water H2O is stronger than either H2 or O2 (see Figure 33). It sticks around on Earth for 4 billion years without any mean of reproduction. |
 |
|
Figure 34 Vesicle |
The capsid of the virus is a protein shell in various forms as shown in Figure 19a. This special coating is made for penetrating the membrane of the host. It may also contain a lipid envelope. The |
 |
|
Figure 35 Minimum Energy |
 |
|
Figure 36 Pre-requisites for |
 Continuation.
Continuation.
 |
 |
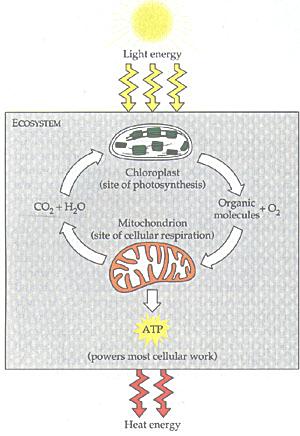
Sun light is captured by chloroplasts to make glucose, which in turn is used by the mitochondria to produce ATP (Figure 08b). The ATP Synthase is ultimately responsible for the chemical process. The chemical formula on the (peptide)n to (peptide)n+1 reaction (and the ATP to ADP, AMP formulas above) actually require a H+ ion on the right-hand-side to make them balanced. It is the concentration gradient of such ions across the cell membrane to complete the conversion from ADP to ATP. This ubiquitous molecule is "used to build complex molecules, contract muscles (including heart beat), generate electricity in nerves, ...". Figure 08c is a cartoon to show the role of ATPs in life. |
Figure 08b Eco-cycle |
Figure 08c Food Cycle |
See the review article "ATP: The Perfect Energy
Currency for the Cell" for detail. |

| The total quantity (number) of ATP in the human body is about 0.1 mole (~ 6x1022). The energy used daily by an adult requires the hydrolysis of 200 to 300 moles of ATP. This means that each ATP molecule has to be recycled 2000 to 3000 times during the day. ATP cannot be stored and so its synthesis has to closely follow its consumption. ATP is formed as it is needed, primarily by oxidative processes in the mitochondria. ATP is not excessively unstable, but it is designed so that its hydrolysis is slow in the absence of a catalyst. This insures that its stored energy is released only in the presence of the appropriate enzyme (to |
Figure 08d ATP in Action [view large image] |
lower the potential barrier) as shown in Figure 08d. Thus, life is an non-equilibrium process (see Figure 22); it ceases to exist once the replenishment of ATP fails to come through. |
 4H+ + 4e- + O2 ----- (1) .
4H+ + 4e- + O2 ----- (1) . |
|
Figure 08e ATP Charging Process [view large image] |
And so it turns the system into a state of thermodynamic non-equilibrium. |
 C6H12O6 + 6O2.
C6H12O6 + 6O2. |
 |
|
Figure 08f Photosynthesis |
Figure 08g ATP Charging Sites [view large image] |
 |
For example, the oxidation of Fe2+ to Fe3+ releases about 0.4 ev in the reaction : 4Fe2+ + 2H2O  4Fe3+ + 4H+ + 4e- + O2 ----- (2), 4Fe3+ + 4H+ + 4e- + O2 ----- (2),which would dispatch the electrons to perform some works and to create an ionic proton H+ gradient in the periplasm (a space between the outer and inner membranes). It is then used to run the ATP synthase to generate ATPs (see Figure 08h and compare the photosynthesis in Eq.(1)). |
Figure 08h ATP by Chemo |
 |
|
Figure 08i Photosynthesis Evolution |
Figure 08i (see details). Finally, the cyanobacteria appeared to possess thylakoid in the cytoplasm as photosynthetic lamellae (see cyanobacteria). |
 |
|
Figure 08j ATP Charging by Glucose |
generate high concentration of H+ ions in the inter membrane space. The gradient enables the ATP-synthase to produce 36 ATPs from each glucose molecule. The ATPs then exit to the cytoplasm for use. |


 ________________________________________
________________________________________ ________________________________________
________________________________________
 ________________________________________
________________________________________


 ,
, 
 click image to see 2025 news on RNA
click image to see 2025 news on RNA
 click image to see "Evolution of the Earth"________________________________________
click image to see "Evolution of the Earth"________________________________________